HELP
-----------------------------------------------------------------------------------------------------------
1 Prediction
To start using PON-IDR, provide UniProt IDs and amino acid substitution information. For protein sequences, there are two ways to input: directly by providing sequences in Fasta format or list of IDs (UniProt). These data can be written or pasted to the boxes in the input forms or uploaded as a file. Both types allow the submission of variants in multiple queries simultaneously. To look for an input example, please click the "Example" text on the input page.
1.1 Input sequence(s)
FASTA sequences have to contain a header line starting with greater than sign (>) followed by amino acids sequence. Amino acids sequence has to start from a new line.
Information for amino acid substitutions has to contain the same header line as the sequence. An amino acid substitution consists of three parts in HGVS format: original amino acid, position, and new amino acid. For example, "A61T" means 61st amino acid A (alanine) is substituted by T (threonine). Use single letter amino acid codes. Each protein sequence can contain multiple amino acid substitutions, each one indicated in a different line.
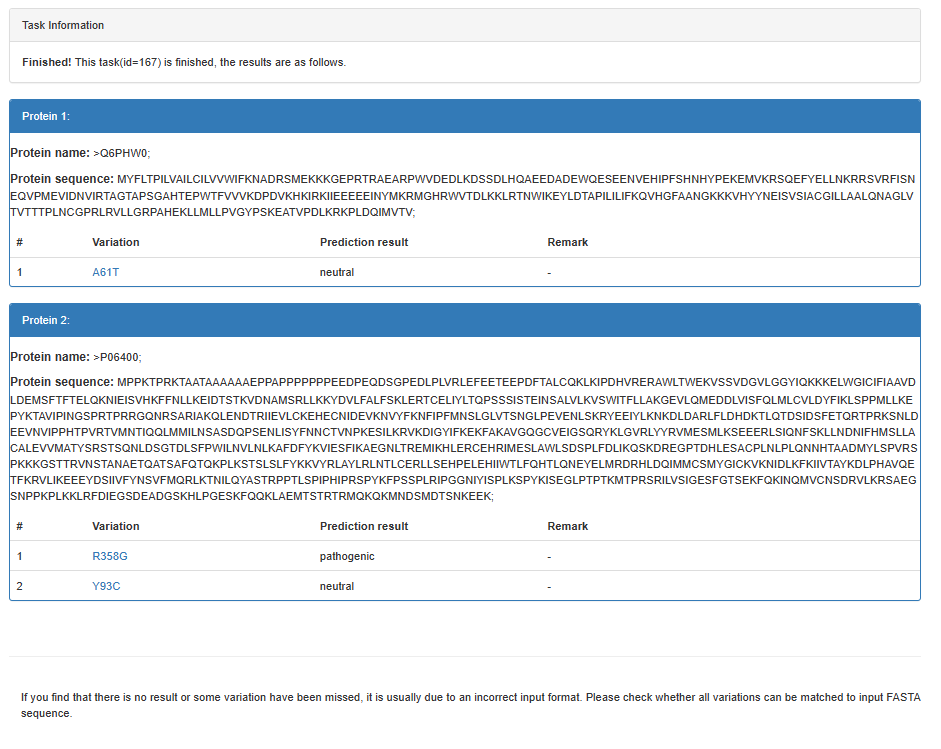
Example
-
FASTA sequence(s)
>P06400
MPPKTPRKTAATAAAAAAEPPAPPPPPPPEEDPEQDSGPEDLPLVRLEFEETEEPDFTALCQKLKIPDHVRERAWLTWEKVSSVDGVLGGYIQKKKELWGICIFIAAVDLDEMSFTFTELQKNIEISVHKFFNLLKEIDTSTKVDNAMSRLLKKYDVLFALFSKLERTCELIYLTQPSSSISTEINSALVLKVSWITFLLAKGEVLQMEDDLVISFQLMLCVLDYFIKLSPPMLLKEPYKTAVIPINGSPRTPRRGQNRSARIAKQLENDTRIIEVLCKEHECNIDEVKNVYFKNFIPFMNSLGLVTSNGLPEVENLSKRYEEIYLKNKDLDARLFLDHDKTLQTDSIDSFETQRTPRKSNLDEEVNVIPPHTPVRTVMNTIQQLMMILNSASDQPSENLISYFNNCTVNPKESILKRVKDIGYIFKEKFAKAVGQGCVEIGSQRYKLGVRLYYRVMESMLKSEEERLSIQNFSKLLNDNIFHMSLLACALEVVMATYSRSTSQNLDSGTDLSFPWILNVLNLKAFDFYKVIESFIKAEGNLTREMIKHLERCEHRIMESLAWLSDSPLFDLIKQSKDREGPTDHLESACPLNLPLQNNHTAADMYLSPVRSPKKKGSTTRVNSTANAETQATSAFQTQKPLKSTSLSLFYKKVYRLAYLRLNTLCERLLSEHPELEHIIWTLFQHTLQNEYELMRDRHLDQIMMCSMYGICKVKNIDLKFKIIVTAYKDLPHAVQETFKRVLIKEEEYDSIIVFYNSVFMQRLKTNILQYASTRPPTLSPIPHIPRSPYKFPSSPLRIPGGNIYISPLKSPYKISEGLPTPTKMTPRSRILVSIGESFGTSEKFQKINQMVCNSDRVLKRSAEGSNPPKPLKKLRFDIEGSDEADGSKHLPGESKFQQKLAEMTSTRTRMQKQKMNDSMDTSNKEEK
-
Amino acid substitution(s)
>P06400
R358G
Y93C
1.2 Input Protein ID
PON-IDR accepts also sequence IDs.The IDs should be preceded by greater than sign (>). After that, provide amino acid substitutions starting from the next line. All the variants in a protein in a single list. After that, details for another sequence can be provided. Substitutions are provided in the HGVS format.
Example
-
ID(s) and amino acid substitution(s)
>P06400
R358G
Y93C
-----------------------------------------------------------------------------------------------------------
2 Prediction result
-----------------------------------------------------------------------------------------------------------
3 Data sets
| Dataset | pathogenic | benign | total |
|---|---|---|---|
| Training dataset | 544 | 464 | 1008 |
| Blind Test dataset | 50 | 50 | 100 |
| noIDR of PON-ALL test set | 2080 | 2267 | 4347 |
| noIDR of ClinVar test set | 7167 | 2089 | 9976 |
The dataset can be download from here.
Training set
Blind test set
-----------------------------------------------------------------------------------------------------------
4 Contact
If you have any problems, please contact Wei Wang (20215227100@stu.suda.edu.cn).